


Selected recent publications and interesting “behind the scenes” stories
We're going to start using this space to post our latest BioRxiv preprints (marked in purple). It takes so long to go through the journal system. Might as well post them here in the meantime.
93) Rua, A.J., & Alexandrescu A. T. (2026) “Solution structure of the novel CH-domain zinc finger from the puberty regulator makorin-3" , BioRxiv preprint!
https://www.biorxiv.org/content/10.64898/2025.12.01.691648v1
Work on a novel zinc finger from a protein that is a repressor of puberty.
92) Jerolamon, J., Alder, N.N., & Alexandrescu, A.T. (2026) “NMR assignments of the human homodimeric mitochondrial ATP synthase inhibitor IF1", Biomolecular NMR Assignments 20, 6.
https://doi.org/10.1007/s12104-025-10257-y
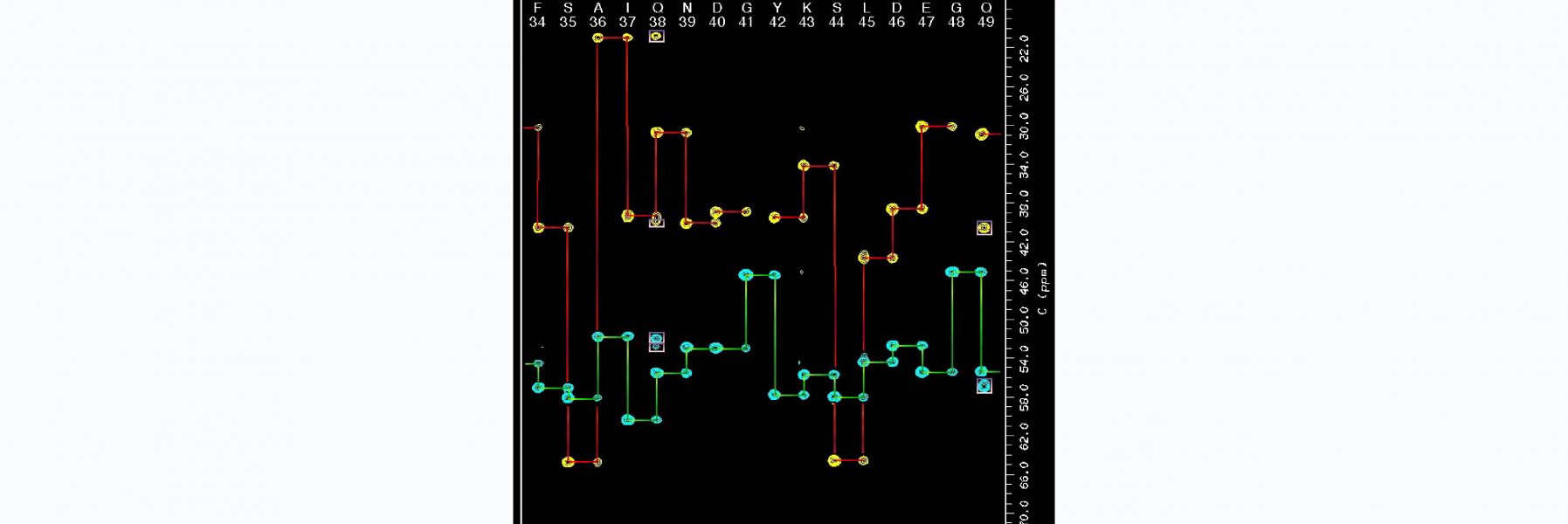
Collaboration with Julia Jerolamon from the Alder lab on a difficult but interesting coiled coil that is a natural inhibitor of ATP synthase. Julia's first foray into NMR!
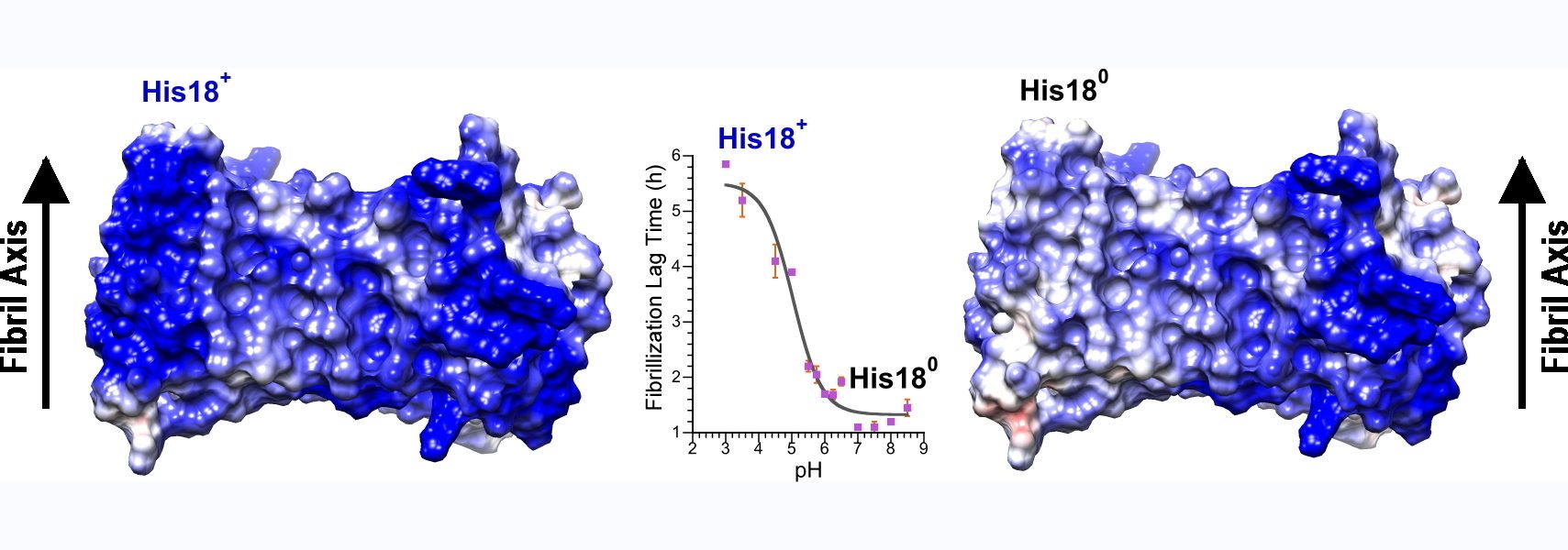
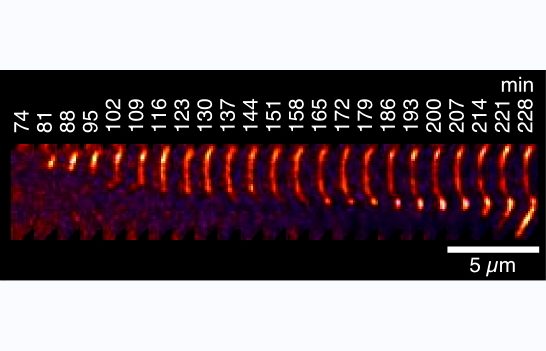
91) Harris, R.E., Rua, A.J. & Alexandrescu, A.T. (2025) “Zinc-induced folding and solution structure of the eponymous novel zinc finger from the ZC4H2 protein”, Biomolecules 15, 1091.
https://doi.org/10.3390/biom15081091
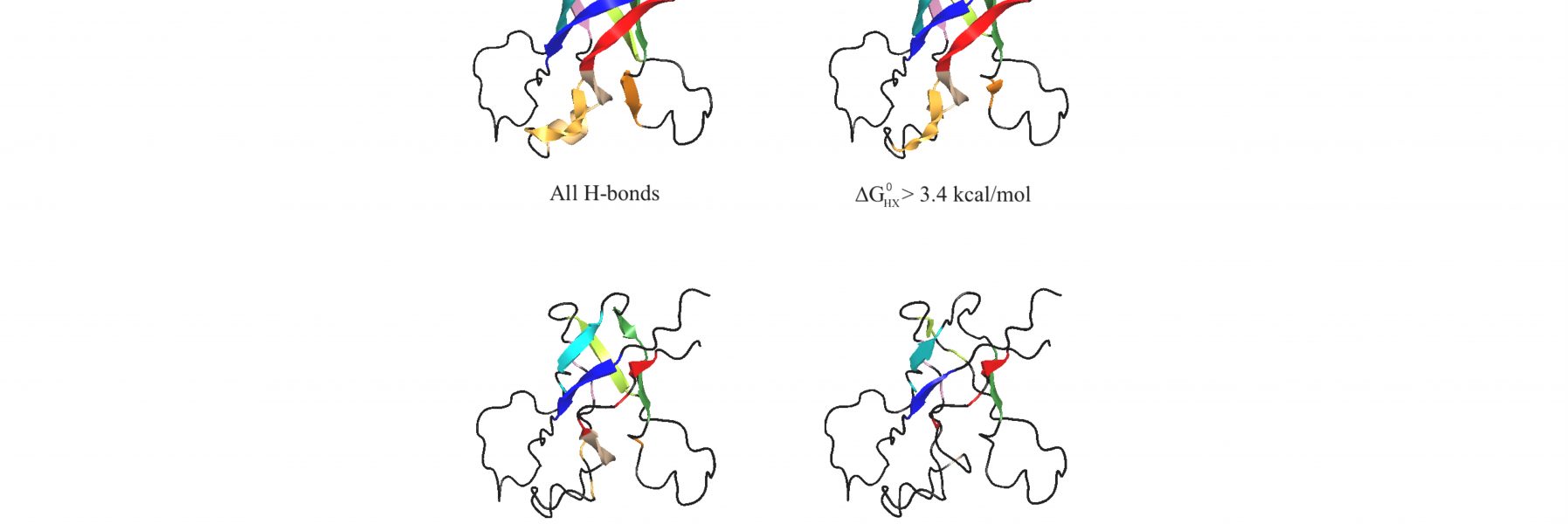
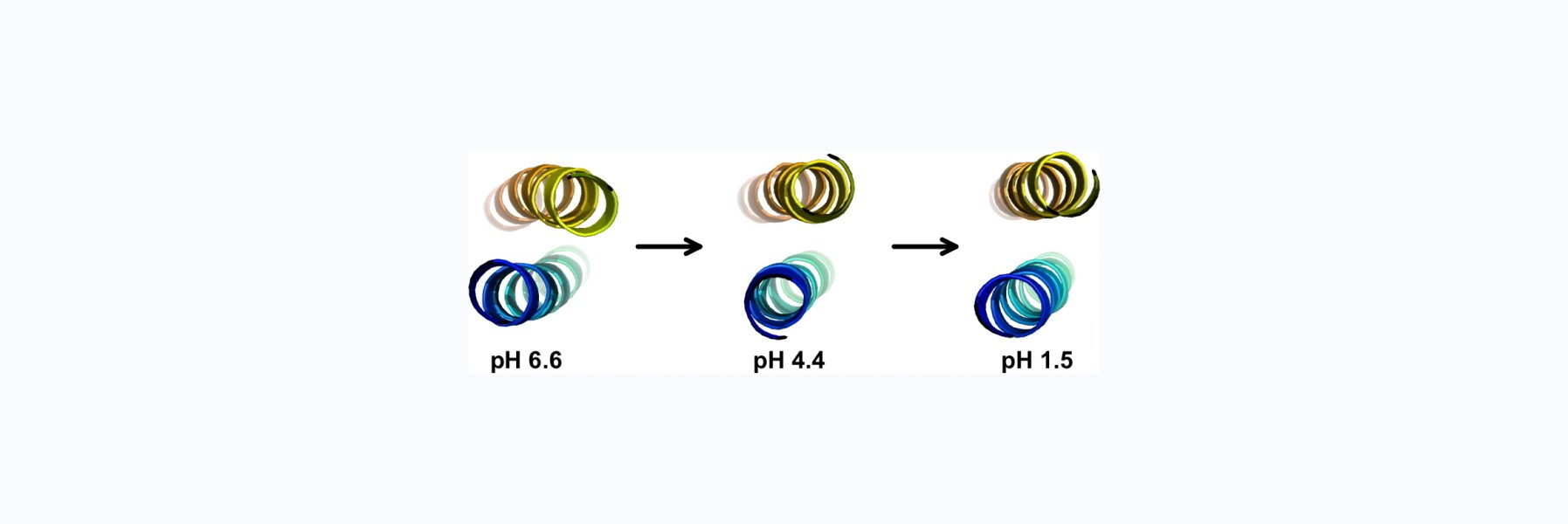
ZC4H2 is a protein consisting of a coiled coil, and a single putative zinc finger with four cysteines and two histidines from which the protein gets its name. The zinc finger is a unique sequence family with no representative structures. Mutations in ZC4H2 cause X-linked congenital developmental diseases with neurological and musculoskeletal dysfunction called ZARD (ZC4H2-Associated Rare Disorders). About ten of the currently known ZARD mutations are in the zinc finger, suggesting it is important for the function of the protein in neurodevelopment. The precise function of the ZC4H2 protein is unknown, and it hasn't even been established if the zinc finger, for which the protein is named, is folded. AlphaFold doesn't give a very clear answer. We therefore undertook biophysical studies of the folding and metal binding of the zinc finger and determined its solution structure. We hope our work will provide a framework for determining the function of Zc4H2 and draw more attention to this interesting gene/protein.
Andrei T. Alexandrescu
| Phone: | 860-486-4414 |
|---|---|
| E-mail: | andrei.alexandrescu@uconn.edu |
| Address: | 91 North Eagleville Road, Unit 3125 Storrs, CT 06269-3125 Biology/Physics Building 209 |