Selected recent publications and interesting “behind the scenes” stories
We're going to start using this space to post our latest BioRxiv preprints (marked in purple). It takes so long to go through the journal system. Might as well post them here in the meantime.
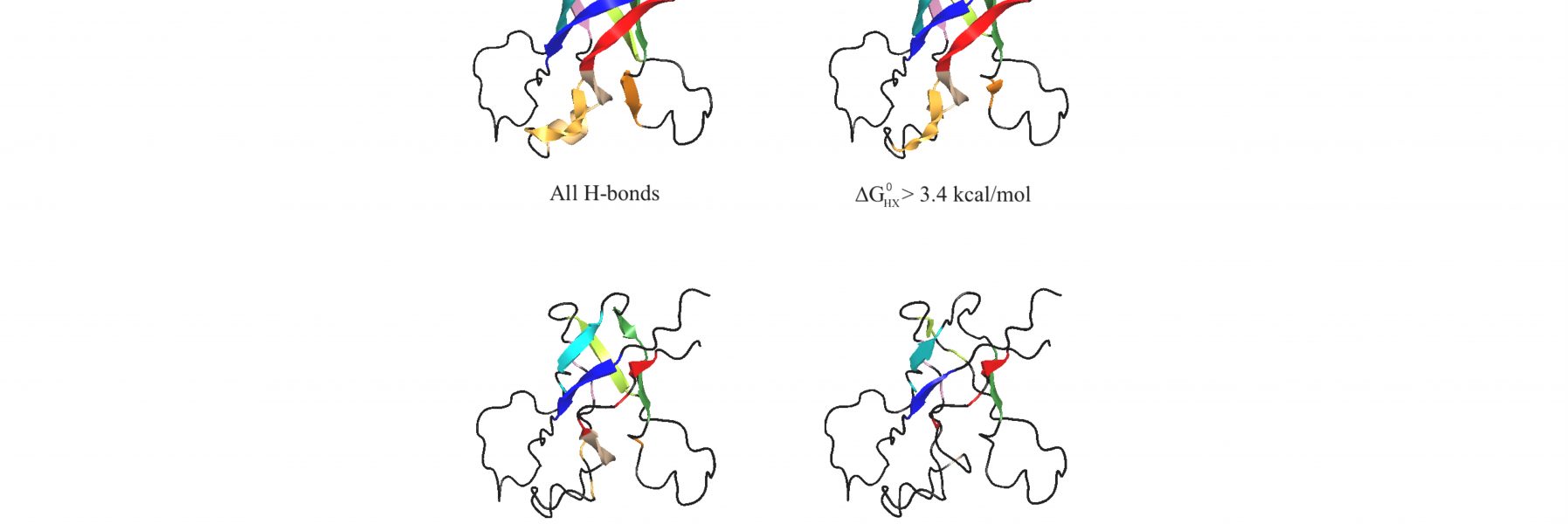
94) Alexandrescu A. T., Rua, A.J., Shah, S., Fairchild, D., & Bezsonova, I. (2026) “High-pH NMR to identify macromolecular H-bonds and foldons" , BioRxiv preprint!
https://www.biorxiv.org/content/10.64898/2026.02.28.708709
Sameena Shah's rotation project, that over a year and a half morphed into a method to study H-bonds in protein structures and unfolding intermediates.
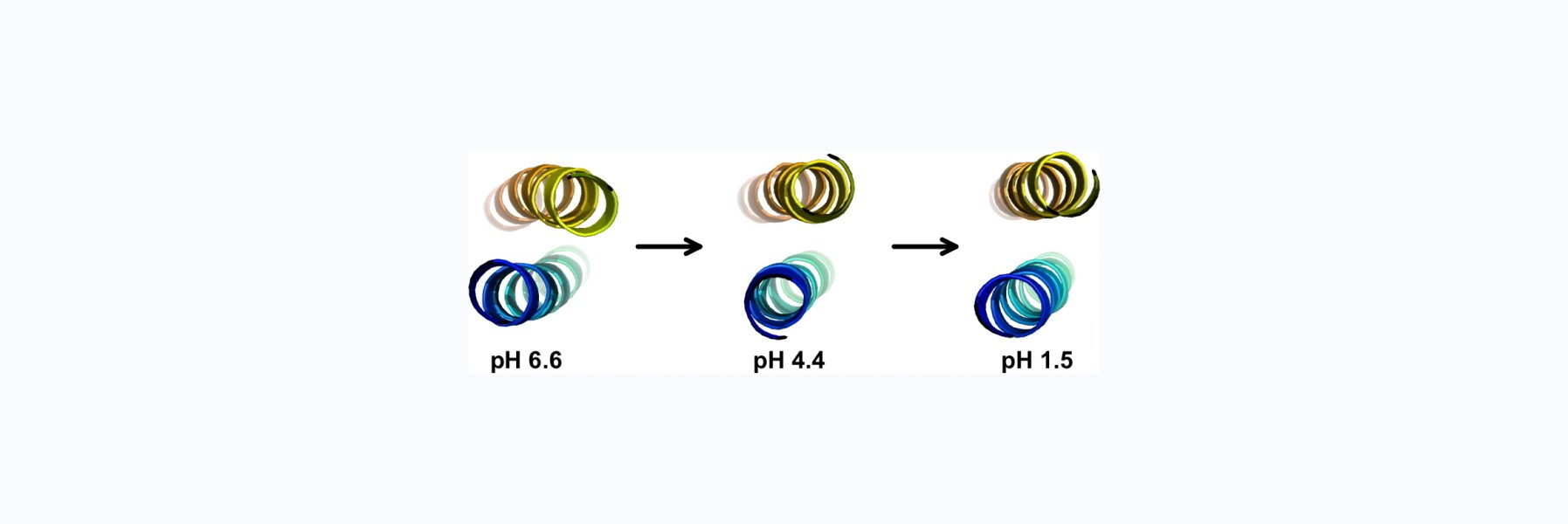
93) Rua, A.J., & Alexandrescu A. T. (2026) “Solution structure of the novel CH-domain zinc finger from the puberty regulator makorin-3", BBRC, in press, coming soon!
BioRxiv preprint: https://www.biorxiv.org/content/10.64898/2025.12.01.691648v1
Work on a novel zinc finger from a protein that is a repressor of puberty.
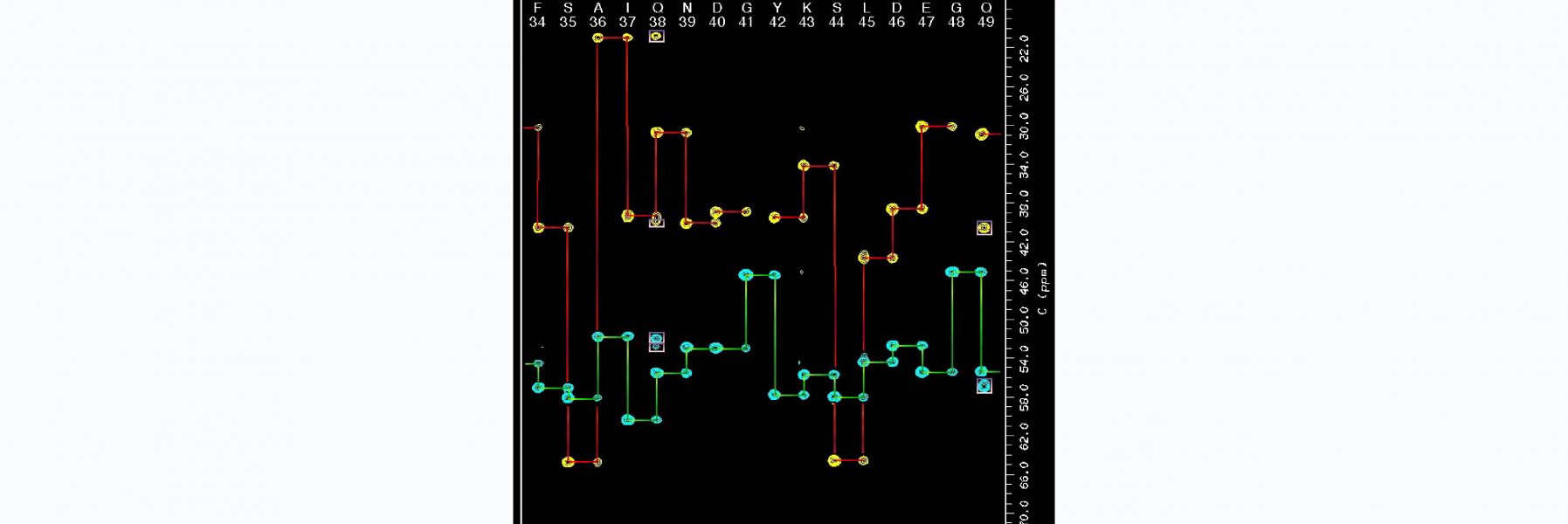
92) Jerolamon, J., Alder, N.N., & Alexandrescu, A.T. (2026) “NMR assignments of the human homodimeric mitochondrial ATP synthase inhibitor IF1", Biomolecular NMR Assignments 20, 6.
https://doi.org/10.1007/s12104-025-10257-y
Collaboration with Julia Jerolamon from the Alder lab on a difficult but interesting coiled coil that is a natural inhibitor of ATP synthase. Julia's first foray into NMR!
Andrei T. Alexandrescu
| Phone: | 860-486-4414 |
|---|---|
| E-mail: | andrei.alexandrescu@uconn.edu |
| Address: | 91 North Eagleville Road, Unit 3125 Storrs, CT 06269-3125 Biology/Physics Building 209 |